The Role of Spatial Biology in Immunotherapy and Cancer Treatment

ONCOLife |
5 April 2024
Dr. Arutha Kulasinghe, from the University of Queensland’s Clinicalomics Lab, discusses the transformative role of spatial biology in cancer research in an exclusive interview for ONCOLife. By exploring the microenvironment of tumors and identifying unique biomarkers, his work aims to personalize immunotherapy treatments, promising improved outcomes for patients by distinguishing between likely responders and non-responders to such therapies.
Dr. Arutha Kulasinghe, highlights the potential of spatial biology in identifying responders and non-responders to immunotherapy, underscoring its role in advancing personalized medicine. With groundbreaking work on biomarker discovery and the development of companion diagnostic assays, Dr. Kulasinghe’s research paves the way for more effective cancer treatments.
Click to view the PDF version: Pg 12-15.
Exclusive Interview with Dr. Arutha Kulasinghe on Spatial Biology
Could you explain what spatial biology is and how it currently plays a role in cancer research and treatment?
Dr. Kulasinghe: Spatial biology is a relatively new field that emerged around four or five years ago. Traditionally, pathologists have utilized spatial approaches in histology, focusing on the distances between cells and the location of tumors within tissues. We integrate this spatial information to deepen our understanding of the tumor microenvironment. This field, in its infancy about five or six years ago, began with the investigation of regions of interest or limited plex-based profiling of tissues at the transcriptome level, along with targeted panels.
Subsequently, we have pursued low plex protein validation of these targets. However, in the past couple of years, high-plex protein analysis has
expanded, offering a new discovery lens for biomarker identification. This allows us to understand cellular communication within and across tissues.
We’re not only looking at where tumor cells are located but also examining subsets of tumor cells, immune cells, and interactions at the tumor microenvironment interface, among others. This new perspective on biology aids in identifying reasons some patients respond well to current therapies while others do not.
Unraveling Tumor Microenvironments
With the high percentage of patients not responding to immunotherapies, how does spatial biology help in distinguishing potential responders from non-responders?
Dr. Kulasinghe: Immunotherapy costs about $200,000 per patient per year. However, it is effective in only about 15 to 30% of patients, which means it does not work for everyone. Our goal is to identify the most suitable immunotherapy for the right patient. For the 70% of patients who do not respond to immunotherapy, we aim to understand what is happening in the tumor biology. This could indicate a need for combination therapy or strategies to attract the right types of immune cells, ensuring the tumor is recognized by the immune system. This represents an unmet clinical need.
Currently, we lack effective methods to accurately identify responders and non-responders. While there are biomarkers in use, such as PD-L1 expression, tumor mutation burden, and microsatellite instability, these do not adequately capture the dynamic changes occurring in the tumor microenvironment.
Spatial biology is enabling us to deeply understand the underlying biology of the individual cells within the tumor and the immune compartment. We’re exploring how these cells interact within the tissue, hoping this will offer new insights into why some patients respond to immunotherapy whi-
le others do not.
How critical is biomarker discovery in advancing our understanding of disease mechanisms, particularly in cancer? Can you discuss the limitations of current diagnostic tests, in predicting immunotherapy response?
Dr. Kulasinghe: Current biomarkers for predicting immunotherapy response primarily focus on single-marker expression on tumor cells, such as PD-L1 on tumor cells and PD-1 on immune cells. The expression levels of these markers can vary widely, ranging from 0% to 100%, with levels like 10% positive, 50% positive, and even 100% positive.
However, this range of expression does not necessarily correlate with the actual res-ponse to immunotherapy, highlighting a limitation. PD-L1 is a dynamically changing biomarker that cannot be fully captured in a single snapshot of the tissue.
Other important biomarkers include tumor mutation burden, which looks at the number of new antigens expressed that the immune system can recognize as foreign, thereby driving an anti-tumor immune response.
Additionally, tumors with high microsatellite instability (MSI-high) generally respond well to treatment. These biomarkers are linked to a high new antigen load, indicating that such tumors typically fare better. However, the challenge lies in the fact that we do not have reliable biomarkers to predict response accurately.
We are unable to differentiate responders from non-responders effectively. Considering the high cost of these drugs -$200,000 per patient per year - and their potential immune-related adverse effects, identifying the right patient for treatment is crucial. This underscores the need for more comprehensive biomarkers that can capture the tumor microenvironment more accurately, thereby predicting response or resistance to therapy.
Your studies in head and neck squamous cell carcinoma and non-small cell lung carcinoma have shed light on the tumor microenvironment. What have been the most surprising findings from this work?
Dr. Kulasinghe: What we’ve discovered is that glucose, or sugar, has a particular affinity for cancer cells. When you undergo an FDG PET scan of your entire body, you’re essentially searching for areas of high glucose uptake. FDG serves as a tracer for glucose within the body. Thus, if you have breast, lung, or prostate cancer, the affected organ—be it the breast, lung, or prostate—will illuminate during an FDG tracer PET scan, indicating macro-level glucose uptake.
In our research, particularly with head and neck and lung cancers, we’ve identified that glucose uptake isn’t uniform across the entire tumor. Instead, there are specific pockets within the tumor that exhibit significantly higher glucose uptake. These areas of intense activity are not found throughout the whole tissue but are highly active in certain regions. We have found that these metabolically active tumor cells, which demonstrate increased glucose uptake, are indicative of resistance to therapy. In our studies, increased glucose uptake in a tumor correlates with a poorer response to immunotherapy.
Glucose appears to be a marker associated with resistance, a phenomenon that ties back to Warburg effect, which is characterized by increased glucose uptake in cancer cells. This leads us to believe that it plays a role in driving a resistant phenotype in tumors.
Considering potential of spatial biology, how do you see its role evolving in the development of companion diagnostics for cancer treatments?
Dr. Kulasinghe: Right now, there is a companion diagnostic assay developed by Macrophone Therapeutics that utilizes a spatial score predictive of a patient’s benefit from a DNA damage drug. This represents one of the first spatial scores capable of identifying patients likely to benefit from the therapy, providing a roadmap for application in other tumor types.
Their objective is to profile a sufficient number of tumors in a retrospective study. This will enable us to identify the biomarkers defining our response or resistance groups and then develop these findings into a companion diagnostic assay using a similar approach. I believe companion diagnostics are poised to become a significant aspect of spatial biology. However, the most impactful developments will probably emerge from the field of spatial proteomics.
What do you see as the biggest challenges and opportunities in integrating spatial biology into routine clinical practice for cancer treatment?
Dr. Kulasinghe: Spatial biology, being a relatively new field, is witnessing a surge of new data. There is a pressing need for assay standardization and harmonization of approaches. It’s crucial that the same assay, when conducted in multiple laboratories using similar tissue types, yields consistent results.
Furthermore, the analytical pipeline used to interpret these results must be robust and reproducible. If a tissue sample were sent to ten different labs, each should report very similar findings regarding the spatial outcome or score. I think that the standardization of workflows and experimental approaches, as well as the analysis, is essential. Moreover, as these datasets become increasingly complex -often comprising hundreds of terabytes -the challenge of analyzing them grows. A cloud computing-based approach might be necessary for this analysis.
Despite having a roadmap for companion diagnostic assays, there’s a lot of groundwork to be done in terms of integrating these advances into clinical practice. We need standardized workflows and field-wide harmonization to advance these assays from the laboratory to clinical settings effectively.
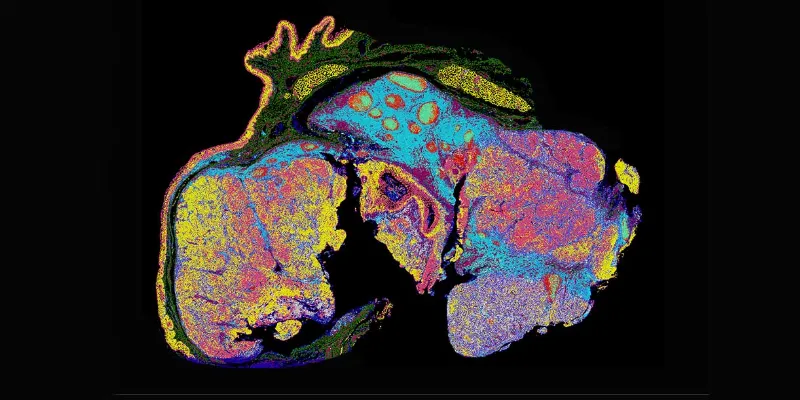
Whole-slide single-cell spatial phenotyping of a human FFPE head and neck squamous cell carcinoma with an ultrahigh-plex antibody panel reveals four distinct tumor regions with varying metabolic and stress profiles. (A) Tumor 1 (T1) has a high degree of apoptotic cells (blue); Tumor 2 (T2) shows a central apoptotic core (blue) surrounded by proliferating cells (light purple); Tumor 3 (T3) shows high expression of markers for the pentose phosphate pathway (peach), fatty acid metabolism (green), and invasion (red); and Tumor 4 (T4) is highly dynamic with cells undergoing glycolysis (light pink), tricarboxylic acid cycle (yellow), and apoptosis (blue). (B) Quantification of the 14 functional phenotypes as listed in the legend shows the predominant pathways within the normal squamous epithelium (SE), normal tonsil tissue (NT), and four tumor regions illustrating the high degree of intra-tumoral heterogeneity and competing microenvironments that may inform the partial response to ICI observed in this case.
Figure is from: Jhaveri, et al. Mapping the Spatial Proteome of Head and Neck Tumors: Key Immune Mediators and Metabolic Determinants in the Tumor Microenvironment. GEN Biotechnology. Published Online:16 Oct 2023. https://doi.org/10.1089/genbio.2023.0029




Comments
No Comments Yet!